dual chain AR
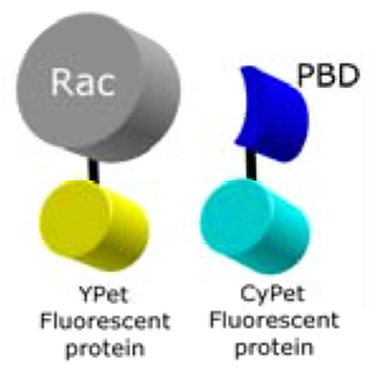
In this Rac1 biosensor an “affinity reagent” derived from Pak1 (PBD) binds specifically to the active form of the GTPase. The biosensor uses FRET as a readout of GTPase binding.
References
Marston, D.J., Vilele, M., Huh, J., Ren, J., Azoitei, M., Glekas, G., *Danuser, G., *Sondek, J., and *Hahn, K.M. Multiplexed GTPase and GEF biosensor imaging enables network connectivity analysis.. Nature Chem. Biol., 16(8): 826-833, 2020. PMC7388658. Online article | Free PMC article
O'Shaugnessy, E.C., Stone, O.J., LaFosse, P.K., Azoitei, M.L., Tsygankov, D., Heddleston, J.M., Legant, W.R., Wittchen, E.S., Burridge, K., Elston, T.C., Betzig, E., Chew, T.L., Adalsteinsson, D., and Hahn, K.M. Software for lattice light-sheet imaging of FRET biosensors, illustrated with a new Rap1 biosensor. J. Cell Biol., 218(9): 3153-3160, 2019. PMC6719445. Online article | Free PMC article
Machacek, M., Hodson, L., Welch, C., Elliot, H., Pertz, O., Nalbant, P., Abell, A., Johnson, G., *Hahn, K.M. and *Danuser, G. Coordination of Rho GTPase activities during cell protrusion. Nature, 461:99-103, 2009. PMC2885353. Online article | Free PMC article
Methods articles
Hodgson, L., Pertz, O., and Hahn, K.M. Design and Optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol, 85:63-81, 2008. [Link downloads PDF] Online article
*Note: this older methods article contains a useful description of biosensor image analysis.
Hodgson, L. Nalbant, P. Shen, F., and Hahn, K. Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation. Methods Enzymol., 406:140-156, 2006. Online article | PDF
*Note: this article is focused on dye-based biosensors, but includes a method to correct for bleaching of any fluorophore.
~ Updated 04/07/2021