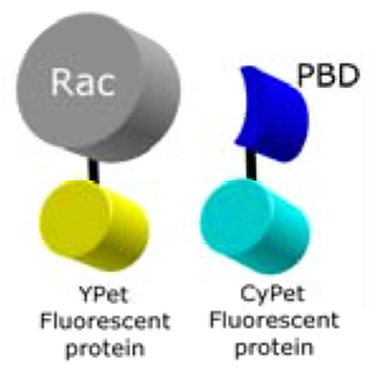
In this Rac1 biosensor an “affinity reagent” derived from Pak1 (PBD) binds specifically to the active form of the GTPase. The biosensor uses FRET as a ‘readout’ of GTPase binding.
Some references
Kraynov, V.S., Chamberlain, C.E., Bokoch, G.M., Schwartz, M. A., Slabaugh, S. and Hahn, K.M. Localized Rac activation dynamics visualized in living cells. Science, 290: 333-337, 2000. Online article | PDF
Machacek, M., Hodson, L., Welch, C., Elliot, H., Pertz, O., Nalbant, P., Abell, A., Johnson, G., *Hahn, K.M. and *Danuser, G. Coordination of Rho GTPase activities during cell protrusion. Nature, 461: 99-103, 2009. PMC2885353. Online article | Free PMC article
Methods articles
Chamberlain, C.E., Kraynov, V. and Hahn, K.M. Imaging spatiotemporal dynamics of Rac activation in vivo with FLAIR. Methods Enzymol., 325: 389-400, 2000. Online article | PDF
Hodgson, L. Nalbant, P. Shen, F., and Hahn, K. Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation. Methods Enzymol., 406:140-156, 2006. Online article | PDF
*Note: this article is focused on dye-based biosensors, but includes a method to correct for bleaching of any fluorophore.
Hodgson, L., Pertz, O., and Hahn, K.M. Design and Optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol, 85:63-81, 2008. [Link downloads PDF] Online article
*Note: this older methods article contains a useful description of biosensor image analysis.
~ Updated 02/17/2021