research
Click for Lab News
Cells are not like machines with a computer chip “brain” that integrates inputs and controls moving parts. They are more like transformer toys, where all parts can undergo extensive rearrangement to perform vastly different functions. The only way to understand such dynamic behavior is to study molecular actions inside living cells.
Our lab focuses on two synergistic areas: 1) developing methods to visualize and control protein activity in live cells and animals, and 2) applying these to address basic questions re spatio-temporal control of signaling. Our biological studies center on the integration of cytoskeletal and adhesion dynamics, and phagocytosis as a model of signaling driven by mechanical force. We are extending our studies to examine the cancer microenvironment in 3D models and in vivo.
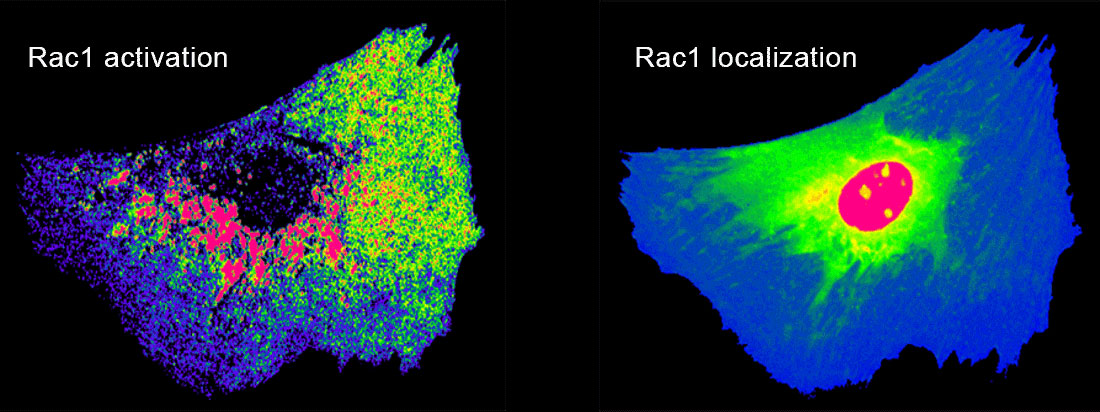
While addressing specific molecules for our biological studies, we produce generally applicable approaches to molecular imaging tools. These include new fluorescent biosensor designs to visualize conformational changes of endogenous proteins and to study the conformation of individual molecules in living cells. We are developing bright dyes that report protein conformational changes, and engineered domains that can be inserted into target proteins to control protein function using either light or small molecules.
In our biological projects, we are currently inducing macrophages to engage geometrically regular objects. Using multiplexed imaging, we simultaneously control and visualize signaling in this system. This enables us to model lines of force and molecular interactions governing target recognition and sheds light on feedback and spatio-temporal control of GTPase networks. In metastatic cells we are asking how GTPases are regulated by multiple GEFs, GDIs, and GAPs with overlapping roles, and how this is affected by the tumor microenvironment.
We are fortunate to work with collaborators whose abilities dovetail with the quantitative data on protein activation dynamics that we can provide. Working with several other investigators, including Gaudenz Danuser, Tim Elston and Richard Superfine, who have been friends and collaborators for years, we can build models of signaling dynamics, apply cutting edge microscopes and use image analysis developed in part for our applications. Through synergy between the diverse expertise we all bring, we accomplish far more than the sum of the parts.
Check here for researchers' descriptions of their ongoing projects
See the "tools" page for information regarding new microscopy and molecular imaging approaches (biosensors, optogenetics, engineered small molecule responses, dyes, image analysis and modeling….)
For their support over the years, we are grateful to the National Institutes of Health, National Science Foundation, American Cancer Society, American Heart Association, Leukemia and Lymphoma Society, Arthritis Foundation, Human Frontiers in Science Program, Department of Defense, NC Biotechnology Center, Deutsche Forschungsgemeinschaft, Autism Speaks, UNC's University Cancer Research Fund, UNC Lineberger Cancer Center, and the UNC Institute for Developmental Disabilities.